Abstract
Background: With the advent of triplet and quadruplet induction therapies, response rates and survival outcomes have improved significantly in multiple myeloma. However, a minority of patients does not achieve deep responses to these anti-myeloma therapies and survival outcomes for these patients remain dismal. The molecular mechanisms of primary therapeutic resistance remain poorly understood and novel therapeutic approaches are needed for these high-risk patients.
Methods: We generated single-cell gene expression and chromatin accessibility profiles at the time of diagnosis from 16 patients with multiple myeloma using the 10X Genomics Multiome platform. Viably frozen CD138+ bone marrow plasma cells were thawed and washed before removing dead cells and isolating nuclei according to the manufacturer's protocols. RNA- and ATAC-seq libraries were constructed per the manufacturer's user guides. GEX and ATAC libraries were sequenced separately on an Illumina HiSeq 4000 instrument before demultiplexing, alignment to the reference genome, and post-alignment quality control. The Cell Ranger pipelines with default parameters were used to trim reads, align, count unique molecular identifiers, and call cells, transcripts, and peaks. Downstream processing was carried out using Seurat and Signac. Doublets were identified by DoubletFinder and batch effects were corrected using Harmony. After quality control for feature counts and mitochondrial gene expression, there were a total of 16188 single cells: 6139 cells from 6 patients resistant to quadruplet induction (Dara-Ixa-Rd), 4841 cells from 4 patients resistant to triplet induction (VRd), and 5208 cells from 6 patients responding to quadruplet / triplet induction. Primary resistance was defined as not achieving at least a very good partial response (VGPR+ after a minimum of 4 cycles of therapy). Expression of the up-regulated resistance genes in single cells was measured by calculating a module score, with higher numbers indicating higher expression (Tirosh et al. Science 2016;352(6282):189-196). We used a score cut-off of 0.200 to define increased expression of resistance genes.
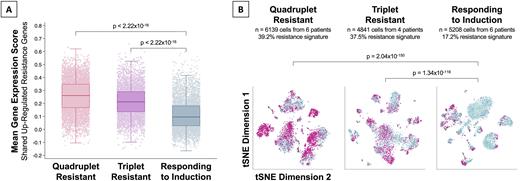
Results: Half of the patients were female, the median age at diagnosis was 61 years (42-80), and half of them had high-risk cytogenetics. There was differential gene expression between cells from quadruplet-resistant and -responsive patients (1272 genes, FDR<0.05). Likewise, there was differential gene expression triplet-resistant and -responsive patients (1354 genes, FDR<0.05). There were 212 genes up- and 222 down-regulated in both comparisons (shared resistance genes), an overlap unlikely to have arisen by chance (p<2.22x10-16). Functional annotation of these shared differentially expressed genes revealed up-regulation of GTPase (FDR=4.01x10-4), kinase (FDR=4.39x10-3), and transferase (FDR=1.65x10-3) activity in cells from resistant patients. Conversely, there was down-regulation of genes involved in ubiquitin (FDR=6.63x10-13) and ubiquitin-like protein ligase binding (FDR=2.50x10-12) as well as unfolded protein binding (FDR=1.21x10-10). Expectedly, the mean expression of the shared up-regulated resistance genes was higher among quadruplet- (score=0.261) and triplet-resistant patients (score=0.212) compared to responding patients (score=0.096, Figure 1A). This difference in gene expression was driven by larger subpopulations of cells with increased expression of resistance genes: Quadruplet-resistant (39.2% of cells) and triplet-resistant (37.5% of cells), compared to patients responding to induction therapy (17.2% of cells, Figure 1B). The subpopulations of cells with increased expression of resistance genes were characterized by increased accessibility in Spi-B (FDR=2.55x10-2) and RAR-γ (FDR=1.86x10-3) binding sites. The shared up-regulated genes included known drug resistance genes such as IKZF1 and viable therapeutic targets including CCND3, RASA1, AKT3, CDK8, and ATM.
Conclusions: Single-cell interrogation of the transcriptome and epigenome of patients with primary resistant multiple myeloma suggests an overexpression of actionable drug targets and increased cell proliferation in cell subpopulations. There is biological rationale for the investigation of novel individualized therapeutic approaches like IKZF1 degradation and targeted kinase inhibition in this patient population.
Disclosures
Kumar:AbbVie,: Membership on an entity's Board of Directors or advisory committees, Research Funding; Celgene: Membership on an entity's Board of Directors or advisory committees, Research Funding; Janssen,: Membership on an entity's Board of Directors or advisory committees, Research Funding; Takeda,: Membership on an entity's Board of Directors or advisory committees, Research Funding; Adaptive,: Membership on an entity's Board of Directors or advisory committees, Research Funding; KITE,: Research Funding; MedImmune/Astra Zeneca,: Membership on an entity's Board of Directors or advisory committees, Research Funding; Merck,: Research Funding; Novartis,: Research Funding; Roche: Research Funding; Sanofi: Research Funding; Oncopeptides: Other: Independent review committee.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal